
Contact
Biography
Chiara is originally from Messina in Sicily, Italy. She obtained her MSci in Pharmaceutical Chemistry and Technology from the University of Messina in 2013. Her research project concerned the development of novel iron chelators as potential drugs for the treatment of Parkinson’s disease under the joint supervision of Professor Robert Hider (Kings College London) and the late Professor Giovanni Romeo (University of Messina).
In 2014 Chiara joined Professor Alessio Ciulli and Professor Dario Alessi labs at University of Dundee to conduct her PhD project on the development of Homo-PROTACs, bivalent small-molecule dimerizers as a strategy to induce self-degradation of E3 ubiquitin ligases. Her work culminated in the discovery of CM11, a homo-PROTAC for the self-degradation of von Hippel-Lindau (VHL). CM11 is now commercially available for the scientific community to use to as chemical tool to rapidly knockdown VHL and is also listed on the Chemicalprobe.org Portal.
Following the award of her PhD in Chemical Sciences in 2017, and a short post-doctoral research in Dundee, in January 2018 Chiara joined the research groups of Professor Chris Schofield and Professor Akane Kawamura at University of Oxford to investigate new molecular mechanisms of epigenetic processes. In February 2020, Chiara was awarded a BBSRC Discovery Fellowship and moved to Newcastle University shortly after, where she started here independent career working on protein post-translational modifications.
Chiara has started her research group in the MRC Protein Phosphorylation and Ubiquitination Unit in January 2023.
Research
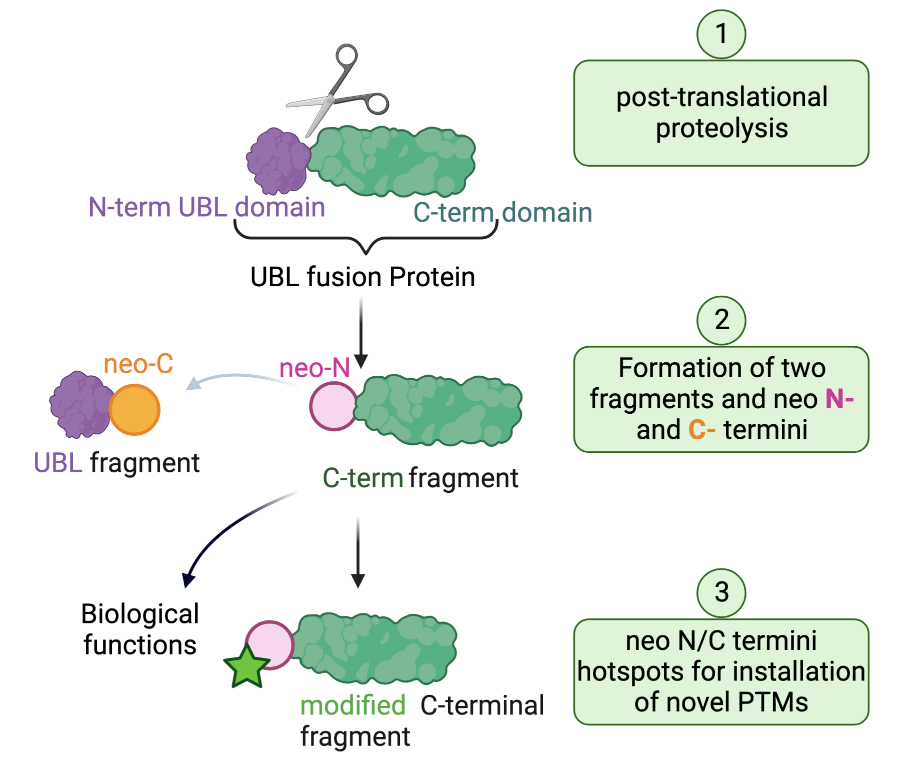
“Cleave-to-Modify”: Illuminating the “dark proteome” with novel chemical biology tools
Variations in protein structure at the level of amino acid sequence and modifications allow for the biological complexity observed in living organisms; a complexity that far exceeds that of the approximately 20,000 genes in the human genome that encode for proteins. Post-translational modifications (PTMs) add to the diversity of the proteome through the generation of new and different proteoforms. Amongst the known PTMs, proteolytic cleavage (or proteolysis) is an irreversible modification performed by a family of specialized enzymes termed proteases. It is characterized by the highly precise cleavage of a protein into two or more smaller fragments defined by distinct new ends generated by the protease at the cleavage site, called neo-N and neo-C-termini. These new fragments can be stable in vivo and can exhibit altered biological activity, location, and interactions compared with their precursor protein. However, the mechanisms by which these processes are regulated remain largely mysterious.
An example of proteolytically processed proteins are the members of the ubiquitin-like domain fusion protein family. These undergo proteolytic cleavage as a mean to regulate their stability and physiological role. We have recently discovered that the generated neo-termini serve as “hot spots” for unprecedented PTMs, a mechanism we refer to as “cleave to modify”. However, not much is known about ubiquitin-like domain fusion proteins, the protease enzymes involved in their processing and the biological events and modifications that are actioned as a result. The long-term objective of the Maniaci Lab is to develop tailor-made tools to illuminate and understand the events that lead to the proteolysis of the “dark proteome” of ubiquitin-like domain fusion proteins and their modifications.
Current research focus:
- Unravel the key players involved in the “cleave to modify” mechanism
- Identify and characterise the PTMs involved and their functional consequences
- Illuminate the biology that is actioned as a result of these events
Often regarded as a mere mechanism for protein destruction, proteolytic cleavage has instead a much more widespread regulatory role in fundamental cellular processes like cellular growth, differentiation, apoptosis, and survival. Consequently, misregulation of proteolytic activities is implicated in many severe acute and chronic human diseases. For these reasons, proteases are considered as attractive drug targets while their cleavage products may provide specific disease biomarkers. Therefore, understanding the proteolytic mechanism and the identity, role and repertoire of the substrates and proteolytic products is not only of fundamental interest but might also reveal invaluable information on the neo-biology of reactivities and recognition, underpinning an unexplored layer of complexity in the hierarchy of cell regulation. This knowledge could therefore be used to identify novel disease markers and allow for the development of specialized therapeutics that target only specific proteoforms.
Values
We are committed to establish a supporting environment where lab members can think freely, express themselves openly, and exchange ideas with mutual respect to enable creative thinking and innovation. This commitment will promote an equal, inclusive, and diverse community to drive scientific progress. We welcome applicants from underrepresented and disadvantaged backgrounds. Please get in touch if you are interested in joining the team.
Selected Publications
- Maniaci, C., Hughes, S. J., Testa, A., Chen, W., Lamont, D. J., Rocha, S., Alessi, D.R., Romeo, R. Ciulli, A. Homo-PROTACs: Bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nat Commun 8, 830 (2017). DOI:10.1038/s41467-017-00954-1
- Zoppi, V., Hughes, S. J., Maniaci, C., et al. Iterative design and optimization yields a potent, fast and selective PROTAC for a target-ligase pair considered unproductive to degradation. J. Med. Chem., 2019, 62 (2), 699-726. DOI: 10.1021/acs.jmedchem.8b01413
- Girardini, M.*, Maniaci, C.*, Hughes, S.J*., Testa, A., Ciulli, A. Cereblon vs VHL: Hijacking E3 Ligases Against Each Other Using PROTACs. Bioorganic & Medicinal Chemistry, 27 (2019) 2466–2479. *Joint first authors. DOI:10.1021/acschembio.8b01016
- Imaide, S.; Riching, K.M.; Vetma, V.; Whitworth, C.; Hughes, S.J.; Trainor, N.; Mahan, S. D.; Murphy, N.; Chan, K.-Ho; Testa, A.; Maniaci, C.; Urh, M.; Daniels, D. L.; Ciulli, A. Trivalent PROTACs Enhance Protein Degradation Through Cooperativity and Avidity. Nat. Chem.Biol. 2021, 17 (11),1157-1167. DOI: 10.1038/s41589-021-00878-4.
Teaching
We welcome undergraduate students interested in doing an internship or research stint in the lab. Informal enquiries are welcome.
Stories

Press release
A University of Dundee researcher has been awarded a prestigious fellowship worth £1.5 million to fund her research for the next eight years

News
Chiara Maniaci has become the latest MRC PPU group leader and has now opened her laboratory.